It's a complex question, and in a future post in this series, I'll attempt to identify ideal metaphors we might use to understand this question. For now, I'd like to focus on how the cell utilizes its genes and controls their expression.
This story should probably begin with Barbara McClintock, who won the Nobel Prize in Physiology or Medicine in 1983 for her discovery of mobile genetic elements (you can read her Nobel lecture here). McClintock discovered in the 1940s that certain pieces of the corn plant's genome could switch positions on the chromosome and change the expression of nearby genes, controlling pigmentation and many other traits.
The idea that gene expression could be regulated was revolutionary. Genetics was in its infancy: the structure of DNA had not even been determined, and genes were viewed simply as static determinants of heredity. McClintock perceived the reaction of the scientific community as “hostile” to her work, and in 1951 ceased openly publishing the results of her research for twenty years. Nowadays, the regulation of gene expression is textbook material.
If we simply look at the myriad cell types within an organism or how an organism interacts with its environment, we will see why gene expression has to be regulated.
Take, for example, different cell types like a neuron (nerve or brain cell) or an adipocyte (fat cell). They look and function in completely different ways.
How do cells regulate the expression of their genes?
To answer that question from a conventional perspective, let us turn for now to chapters 4, 6, and 7 of Molecular Biology of the Cell, the definitive guide to mainstream molecular biology. The first author, Bruce Alberts, was President of the U.S. National Academy of the Sciences for twelve years from 1993-2005. The information in this blog post will come from this source except where otherwise stated.
The Genome Is Three-Dimensional
The first thing we must realize in order to understand how our cells regulate the expression of our genes is that the genome is not simply a two-dimensional, linear sequence of its basic building blocks, called nucleotides, but is a complex, dynamic, three-dimensional structure.
In fact, it just has to be three-dimensional. The typical human cell contains two meters of DNA that it must pack into a nucleus measuring about six micrometers in diameter. A micrometer is one millionth of a meter, so this is like packing 24 miles of thread into a tennis ball!
But that misses the point. The three-dimensional structure of DNA is dynamic and highly regulated, and the cell uses it to regulate gene expression in both transient and heritable ways. Thus it is important both in minute-to-minute, day-to-day function, as well as in forms of epigenetic inheritance.
Here is a diagram showing the different levels of organization imposed upon DNA:
The DNA begins as a two-stranded spiraled thread called a double helix, likely a familiar term to most of us who passed high school biology recently enough to have remembered anything from it. But then it is wound around proteins called histones and thereby arranged into structures called nucleosomes that are arranged along the DNA like beads on a string. This beads-on-a-string structure, coated in a number of other non-histone proteins, is called chromatin.
The chromatin is then organized into a number of different layers of folding. The familiar X-shaped chromosome is compacted 10,000-fold, but only occurs during mitosis, the process by which a parent cell divides into two daughter cells. In a non-dividing cell, the chromatin is compacted on average 500-fold.
The importance of histones eluded scientists for decades. The fact that DNA is the vehicle of inheritance was determined in bacteria, which lack histones. Thirty years ago, scientists figured histones were, in the words of the authors of Molecular Biology of the Cell, “relatively uninteresting proteins,” simply aiding in the packing of chromatin but acting as “uninvolved bystanders” in gene expression.
Nothing could be further from the truth.
The mass of histone proteins within chromatin is equal to the mass of DNA. The precise amino acid sequence of histone proteins is so critical to function that organisms as disparate as the cow and the pea — yes, those little green suckers we eat! — differ in sequence less than two percent. That's roughly the sequence difference between chimpanzees and humans for the average gene.
In yeast, nearly every mutation in a histone protein tested has proved lethal. The few mutations that are not lethal cause changes in gene expression and other abnormalities. This is rather remarkable because, according to a 2008 paper, when yeasts are faced with nutritional abundance, 80 percent of their genes can be deleted with no obvious effect at all. Clearly, histones are not “uninvolved bystanders.”
The cell possesses a vast array of enzymes used to modify histones. These include enzymes that chemically modify histones with methyl, acetyl, or phosphate groups, to strengthen or loosen their bonds with the DNA. They also include dozens of different chromatin-remodeling complexes that slide the nucleosome along the DNA and, in conjunction with histone chaperone proteins, can even remove part or all of the nucleosome.
These are all essential to the expression of genes, because the chromatin must be fully unwrapped in order for a gene to be expressed.
The chemical modifications of histones can exist in thousands of different combinations, and are thought to constitute a “histone code.” Cells also insert and remove special “histone variants” into the chromatin as part of this code. In the fruit fly, over 50 proteins have been identified that act as code-reader/code-writer complexes. The code can signal the beginning of DNA replication, the need for DNA repair, or the expression level of a gene.
Some histone modifications are often reversed almost immediately — for example, the sliding or removal of histones during the active expression of a gene — while others may persist for generations, and thus constitute a form of epigenetic inheritance.
While most of the chromatin within the nucleus is packaged as euchromatin, corresponding roughly to the third level of organization shown in the picture above, over ten percent is packaged as heterochromatin, which is dependent on histones, non-histone proteins, and small RNA molecules called interfering RNAs. This is a highly condensed form that silences genes not only within the heterochromatin, but even nearby it.
There are likely to be over ten unique types of heterochromatin that have different magnitudes and types of reversibility. Different heterochromatin patterns are found in different tissues. Thus, heterochromatin likely plays a role in day-to-day function as well as epigenetic inheritance.
Heterochromatin is particularly concentrated in the telomeres located at the ends of chromosomes, and the centromeres that constitute the middle of the “X” shape. The centromere contains a special histone variant and a number of other unique proteins. In humans, there is no DNA sequence that dictates the centromere, so this is another form of epigenetic inheritance. Since a gene's distance from the centromere will influence its level of expression, this is an important form of epigenetic inheritance.
Of course, unraveling the three-dimensional structure is just the first thing that the cell must do to express a gene. Much more must happen at the site of the two-dimensional, linear sequence.
Gene Regulatory Sequences
Genes are associated with regulatory sequences that lie just upstream from them in the DNA sequence, as well as far, far away from them. These interact with regulatory proteins that communicate the needs of the cell and the organism to the cellular machinery that will express the gene.
Some genes require just a handful of regulatory proteins to be expressed and others require hundreds. Humans have about 2,000 different regulatory proteins that are believed to constitute about eight percent of the human genome.
In addition, there are certain proteins that always need to be there. These include RNA polymerase, which actually synthesizes the mRNA transcript, five “general transcription factors” that are actually complexes of 27 total proteins, and a giant complex called Mediator that is composed of 24 individual protein subunits.
Here is a picture of a “gene control region” being activated:
Nevertheless, even from this picture we can see that the activator is actually binding to a location quite far away from the gene, as indicated by the dotted lines in the DNA sequence on the left side. This distant part of the DNA sequence becomes close to the gene by folding of the chromatin. Thus, even in this relatively unraveled state, expressing a gene is a three-dimensional operation.
In fact, the average gene size is 27,000 nucleotides — and as we will see below, only an average of 1,300 of these actually code for the protein — but this elaborate control region can often span 100,000 nucleotides.
The job of the activator proteins is to attract RNA polymerase and the associated transcription factors and Mediator complex, appropriately position them, and chemically modify them so they can get going along the DNA strand. They also must recruit histone modification enzymes, chromatin remodeling complexes, and histone chaperone proteins, so that those “beads on a string” can be plowed through.
There are other gene regulator proteins called “repressors,” and in fact some proteins can act as “activators” in one context and “repressors” in another. As an example, the nuclear receptors for the fat-soluble vitamins A and D can act as repressors in the absence of these vitamins, but as activators in their presence.
In addition to protein activators and repressors, humans also express 400 different micro RNAs (miRNAs) that can form RNA-induced silencing complexes that regulate at least one third of all human genes. These complexes as well as direct addition of methyl groups to cytosine nucleotides within the DNA provide additional mechanisms of epigenetic inheritance.
This large number of genes reflects the exceedingly complex network of controls governing expression of mammalian genes. Each gene is regulated by a set of gene regulatory proteins; each of those proteins is the product of a gene that is in turn regulated by a whole set of other proteins, and so on. Moreover, the regulatory protein molecules are themselves influenced by signals from outside the cell, which can make them active or inactive in a whole variety of ways. Thus, we can view the pattern of gene expression in a cell as the result of a complicated molecular computation that the intracellular gene control network performs in response to information from the cell's surroundings.
The Cell Edits mRNA, Blurring the Definition of “Gene”
As noted above, the average gene is 27,000 nucleotides long, but on average only 1,300 of them code for protein. What's up with the rest?
Here's a picture of two different genes, a small one on the left, and a larger one on the right:
The red stripes are the parts of the gene that code for proteins. These are called exons because they are expressed. The intervening orange sequences are called introns.
Before the mRNA exits the nucleus, the introns must be spliced out. Why, then, do they exist? One explanation given by the authors of Molecular Biology of the Cell is that it would allow exon shuffling during the course of evolution, so that distinct functional domains could be mix-matched in a copy-and-paste manner to generate new proteins.
However, it turns out that not all introns are thrown away. Indeed, the 150 guide RNAs that are involved in the production of the ribosome, the factory where proteins are synthesized, are often encoded by introns. Similar RNA molecules have just recently been discovered that are only produced in the brain, where they are believed to organize the direct chemical modification of mRNA transcripts.
But as we'll see, the most important reason is that splicing allows another level of regulation.
Many genes exhibit alternative splicing patterns, meaning that the cell can make more than one protein from the same mRNA transcript. mRNA transcripts from a full 75% of human genes undergo alternative splicing, often generating dozens of different proteins.
Like the process of transcribing the mRNA in the first place, the cell has activators and repressors with which it can regulate the alternative pathways of mRNA splicing.
In addition, mRNA transcripts from about 1,000 human genes are believed to undergo RNA editing, which involves the chemical modification of some of the nucleotides. For example, we possess enzymes that convert the nucleotide cytosine to uracil, and others that convert adenine to inosine.
This makes it difficult to define what a gene is, and to count the number in the human genome. At one time, it was thought there was a gene for each protein, but this is clearly false. Some have suggested a gene be defined as each unique mRNA, in which case there could be hundreds of thousands. The authors of Molecular Biology of the Cell suggest that a gene be defined as a closely associated cluster of exons that code for a closely related family of proteins, as alternative splicing usually leads to proteins of related function in humans. Very few examples have been identified where radically different proteins are produced from the same sequence, and in these cases this can be considered two distinct genes that overlap in the DNA sequence.
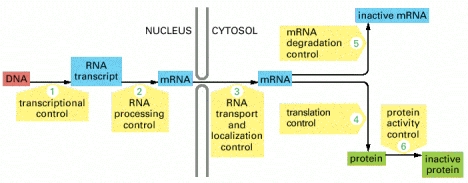
Regulation of mRNA Stability and Translation
The final mRNA transcript doesn't last forever. The cell can regulate how long it lasts, as well as the rate at which it is translated into protein. This begins in the nucleus when the poly-A tail and the poly-A-binding proteins are added, but it continues in the cytosol.
For a nutritional example, the cell has an elegant way of regulating its concentration of iron. There is a sequence of untranslated RNA called an iron response element (IRE) in the mRNA both for ferritin, an iron storage protein, and transferrin, an iron transporter. Ferritin binds iron and thus makes less available for use, whereas transferrin helps the cell take up more iron from the blood.
In ferritin, a single IRE exists in the 5′-untranslated region, near the cap. In transferrin, multiple IRE's exist in the 3′-untranslated region, near the poly-A tail.
When the cellular concentration of free iron gets low, a protein called iron regulatory protein (IRP) loses one of its iron atoms. This causes it to bind to the IREs in the ferritin and transferrin mRNA transcripts.
Because of the different locations of the IREs, IRP-binding causes opposite effects in ferritin and transferrin. It prevents the translation of ferritin protein from its mRNA transcript. By contrast, it prevents the degradation of the transferrin mRNA transcript, causing an increase in the level of mRNA and thus more of the protein to be produced.
Thus, when the iron concentration of the cell gets low, the cell makes more transferrin and less ferritin, both of which increase the cellular concentration of usable iron.
Another way of regulating mRNA degradation is to induce it with little molecules of RNA called interfering RNAs, the same molecules that are sometimes involved in the formation of heterochromatin. There are also a great number of proteins involved in the process of translating the protein, and the cell has a variety of enzymatic networks for controlling these proteins.
Once the protein is produced, there are a great number of ways to regulate its activity, which will be considered in a future post in this series.
Other Forms of Regulation in Viruses and Bacteria
Regulation of gene expression in bacteria is a lot different. Their genomes are much simpler, lacking histones, introns, and true chromosomes, usually regulating lots of genes at once as part of a complex called an operon, and using an expansive list of enzymes to carry it all out, but nevertheless a much smaller list than we use.
Nevertheless, bacteria and even viruses have some sophisticated ways of regulating gene expression that, to date, have not been considered very important in humans and other higher organisms.
Although I learned in genetics class about five years ago that the genetic code has, since the days of Watson and Crick, been considered “non-overlapping,” meaning that each set of three nucleotides constitutes a distinct codon, this is untrue in certain viruses.
Retroviruses, for example, use translational frameshifting to produce their protein shell, called a capsid, and the enzymes reverse transcriptase and integrase that they use to insert their genetic information into their host, all from the same mRNA transcript. They do this by changing the “reading frame,” so that that the codons begin with different nucleotides.
There are cases in bacteria where the bacteria literally restructure their own genome to regulate their genes. For example, Salmonella has a specific 1000-nucleotide DNA sequence that it inverts in order to express one of two different types of flagella proteins. It uses this process to exhibit phase variation.
The bacterium that causes gonorrhea uses a different process called gene conversion to transfer DNA sequences from an unexpressed “library of silent ‘gene cassettes'” to a site in the genome where the genes are expressed, as if playing the “gene cassette” in a “genetic tape player.” This allows it to induce a heritable change in its surface properties and thereby evade an immune attack.
We have already considered organized genome restructuring in humans in the case of antibodies and T cell receptors in the first post in this series. Whether such organized restructuring of the genome has contributed in a greater capacity to who we are on evolutionary and other levels will be considered in a future post.
What Are the Roles of Epigenetics?
So far we've considered histones and associated proteins, RNA-induced silencing, and DNA methylation in epigenetic inheritance. In the last post, I considered protein-folding, which I would consider non-genetic but some would consider epigenetic. What roles do they play?
Certainly, they contribute to cell type. Epigenetic switches and gene expression feedback loops are important in maintaining the structural and functional differences between different types of cells.
They likely also transmit information from the mother's womb and her environment during fetal development, some of which may persist through life.
Do they also contribute to inheritance from one generation to another? That will be the subject of a future post.




Thanks, Chris. Look forward to read more from you.
At any rate your synopsis on "genetics" is indeed marvelous, highly useful and thought provoking. Too many biologists still stick more or less to the plain "central dogma of molecular biology" or other "genetic program" metaphors.
Hey Ned, thanks! I do hope it proves readable to most people. It's a complex topic and takes some effort to fit condense a couple hundred pages of molecular biology text into a readable blog post.
Aaron, very true, although given how much of an influence Paley had on Darwin and given that Darwin used the example of the eye as well, I would say that Paley's eye is Darwin's eye.
Nigel, thanks!
Anonymous, I'll try to answer the 'driver's seat' question later. There is centrality and hierarchy to the governance of the organism, and thus the central nervous system. At the cellular level it is more difficult to identify the locus of intelligent control because the function of information-processing is rather dispersed. But something along the lines of 'what constitutes the brain of the cell?' will be a future post in this series.
Chris
Nice topic. It looks like nobody is in the driver's seat.
Rather a matter of self-organization and emergence in complex systems
As someone who likes to know how stuff works, I am enjoying this series. Great work!
I've often said that the computational complexity of a single cell (even a protazoan) is vastly more complex and dynamic (and non-linear) than the linear operation of even our most advanced computers. This is a marvelous synopsis you provide. William Paley and his example of the eye has got nothing on the level of complexity described at the level of gene expression. It should engender a greater respect for the mechanism of natural selection.
This series is very good Chris. Most people will have to come back to these posts and re-read them, or read them very carefully.
My understanding of evolution is mostly mathematical, from the mathematical foundations that bring together genetics and evolution proper. This work started with Fisher, Haldane and Wright.
This series highlights a number of issues that are very important, and in a way that is accessible without trivializing the topic.